Top Ten Modifications for Nucleotide Synthesis
Top Ten Modifications for Nucleotide Synthesis
We've whittled the list down to ten major modifications and their compounds
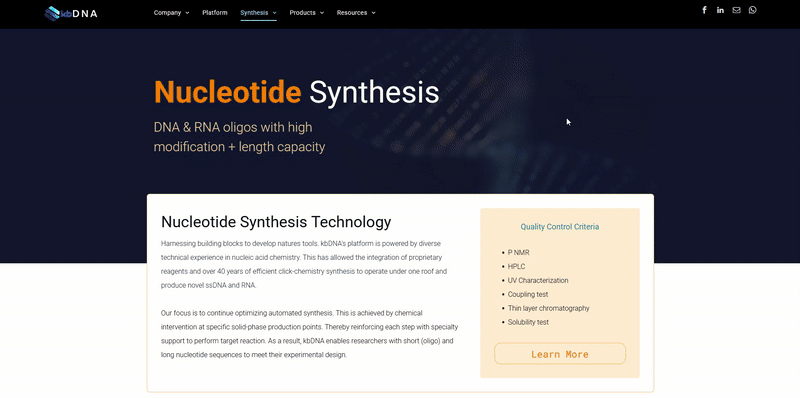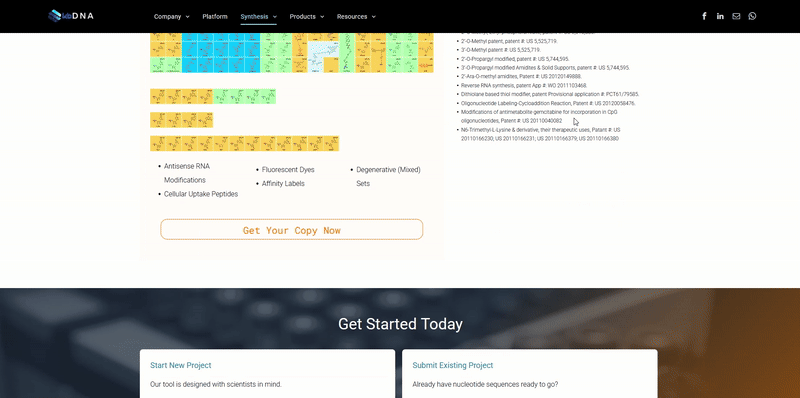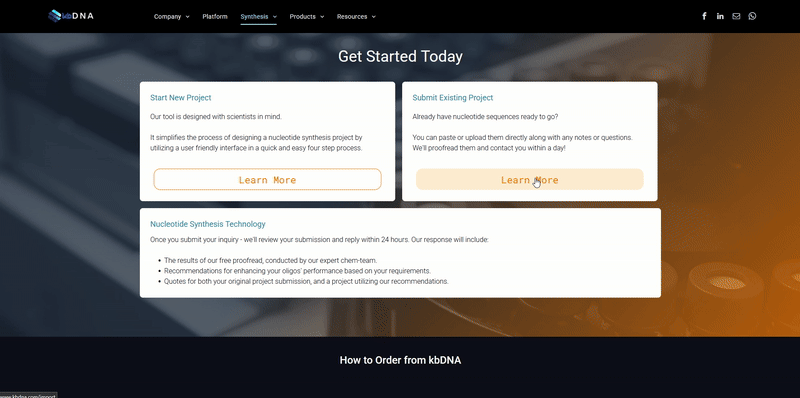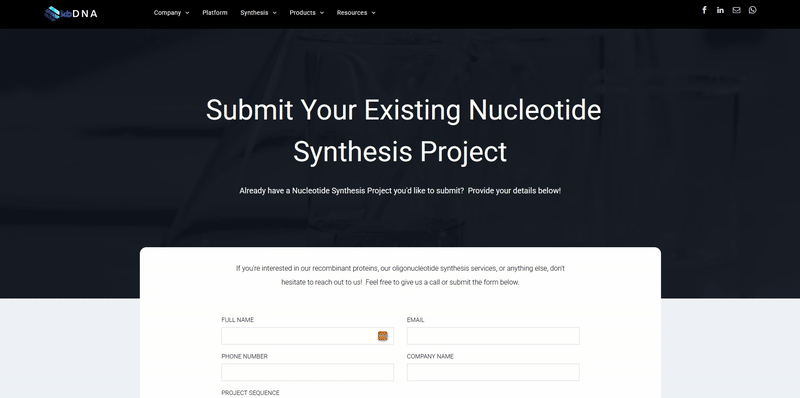
In a previous note , Rethink Modifications; we provide a general understanding for the two main classes of synthesis modifications. We also mentioned how increasingly difficult it has become to keep up with the multitude of list options for each class. With over a hundred elements and endless chemical possibilities, these lists of modification aren’t going to get any shorter. Especially with the growing trends, incorporating nanotechnology (upcoming technical note). We leveraged kb’s competency in this field to help filter out mods weak in principle and outlined the ten major modifications and their differing compounds.
Base Modifications
A base modification focuses on the amino acid base of one or more nucleotides within the nucleic acid sequence. There are three notable modifications to consider. They are as follows:
| 2'-Fluoro | 2'-O-Methyl / 5'-O-Methyl | 2'-O-Propargyl |
|---|---|---|
| Increase Tm, support aptamer selection, and nuclease resistance | Increased Tm of duplexes by 1 - 4°C / block. Provide added stability against hydrolysis and nucleases. | Enzymatic preparation primarily in support of click-based functionality of the Nucleic Acid. The modification allows better constructing of several fluorescent probes to a sequence design. |
| Pyrimidines / Purine |
Sequence Modifications
A sequence modification is any mod that adds or edits features to the nucleic acid sequence. This class offers the highest opportunity for new and different compounds, explaining why most of the modification confusion and errors arise within this category. Many are imitations or slight alterations to similar formulations that result in unnecessary cost and low proof of concept.
We encourage exploring the following seven sequence modifications, prior to making a decision:
| Affinity | Cellular Uptake |
|---|---|
| Minimizing steric hindrance or supporting purification of the biomolecule. Biotin labels, spacers and other chemical compounds allow the use of electronic intervention. | Facilitates the uptake of the compound biomolecule into a cell. Useful in aiding transfection and/or antisense oligos and siRNAs in varying conditions. |
| 3'-Biotin / Biotin (standard) / Biotin-TEG / Cy5.5-AminoC6 | 3' Cholesterol / 5' Cholesterol / 5'-external Cholesterol |
| Degenerate | Fluorescent |
|---|---|
| A combination of two or more different bases at a specific location in a sequence. They can support probe design for strategic hybridization to a known region of an unknown gene. Also known as \""wobbles\"" or \""mixed bases\"". | Chemiluminescent-based labels. They function as probes or dyes to support analysis or aid detection of the biomolecule. |
| 2'-deoxy Degenerate / 2'-Fluoro Degenerate / 2'-O-Methyl Degenerate / 2'-O-Propargyl Degenerate / 2'-ribo Degenerate | 5' ROX Fluorophore / 6-FAM Fluorophore / 6-TAMRA Fluorophore / JOE Fluorophore / ROX Fluorophore |
| Stability | Structure |
|---|---|
| Support different mechanisms different DNA repair mechanisms | Compounds designed to couple with other nucleotides at alternative positions of the ribose than the traditional 5′–3′ direction. This allows the synthesis of more creative sequence designs and concepts such as RNA hairpins, reverse structure, etc. |
| 2'-O-Methyl Inosine / Deoxy Uridine | 2'-5'-RNA / 2',3'-Cyclic Phosphate / L-DNA / L-RNA |
| Terminal Phosphate | |
|---|---|
| Phosphates can be placed on either terminal to block elongation. They aren’t the most optimal for blocking elongation, however their ability to guide enzymatic reaction to form longer oligonucleotides keeps them on the priority mods list. | |
| 3' Terminal Phosphate / 5' Terminal Phosphate / Terminal Diphosphate / Terminal Phosphate / Terminal Triphosphate |
Choose by function…
Ultimately, your intended function provides 90% of a modifications requirements. That is why we encourage spending more time understanding your experimental objective before even considering the different modifications. Once this aspect is understood, it is a matter of identifying and matching biochemical components. kb’s synthesis is focused on proper intervention in the latter, as it is more challenging for scientists who don’t have time to follow the trends of commercial synthesis. Our platform building tool and proofreading services have been able to consistently match the right modification solution to the right function without any fees or added cost. Providing a proper resource has helped many of our end-users avoid errors and unnecessary costs to meet the following functions:
- Substrate for DNA ligase
- Attaching nucleotide to another biomolecule or surface
- Targeted detection
- 3’ blocking or steric hindrance
- Altering hybridization characteristics
- Nuclease degradation concerns
- Novel transfection into cell
Our resources are openly available to all researchers on our
nucleotide synthesis page. Please visit for more information!
References
- Korreck. J. Eur. J. Biochem. 2003, 270, 1625.
- Wilson, C. Curr. Opin . Chem. Biol. 2006, 5487-5502.
- Grimm. D. Adv. Drug, Deliv. Rev. 2009, 61,672-703.
- Lin, C. et. al. Biochemistry 2009, 48, 1663-1674.
- Dubendorff, J. W.; de Hasett, P. L.; Rosendahl, M. S.; Caruthers, M. H. J. Biol. Chem. 1987, 262, 892.
- Brennan, C. A.; Van Cleve, M. D.; Gumpert, R. I. J. Biol. Chem. 1986 261, 7270-7279.