Hsp90 alpha
Recombinant ID:
2260
Request Datasheet
Gene of Interest
Gene Synonyms:
Protein Names:
Accession Data
Organism:
Homo sapiens (Human)
Mass (kDa):
84660
Length (aa):
732
Sequence:
MPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKEDKEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIVTSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKNDKSVKDLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLEGDDDTSRMEEVD
Proteomics (Proteome ID):
Heat shock protein HSP 90-alpha (Heat shock 86 kDa) (HSP 86) (HSP86) (Lipopolysaccharide-associated protein 2) (LAP-2) (LPS-associated protein 2) (Renal carcinoma antigen NY-REN-38)
Proteomics (Chromosome):
UP000005640
Mass Spectrometry:
N/A
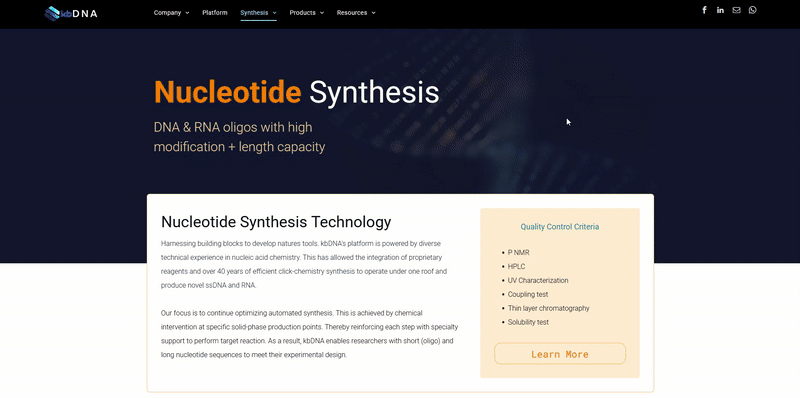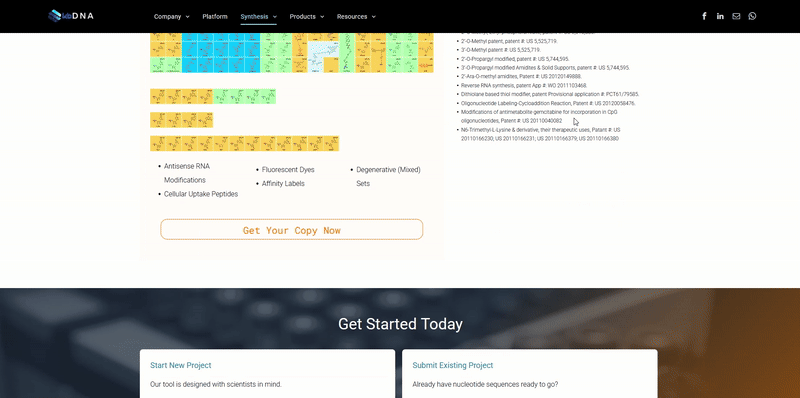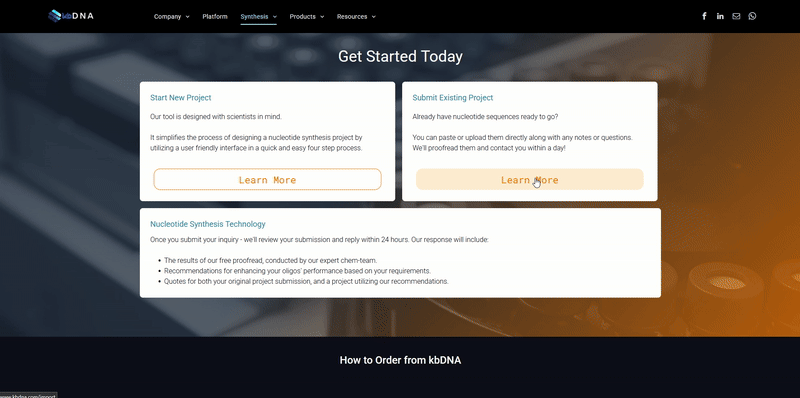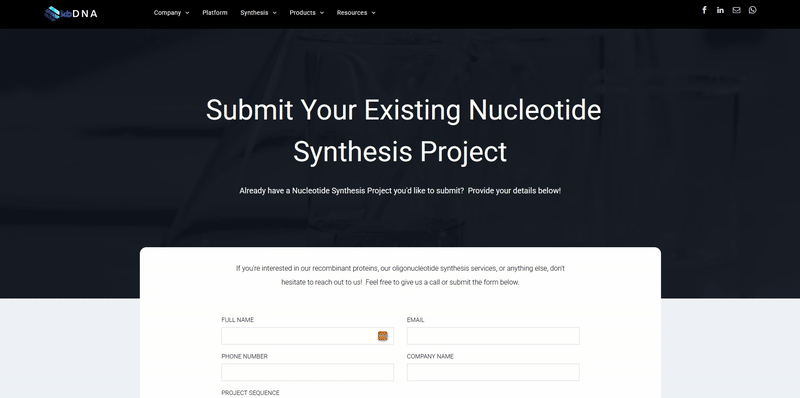
Function [CC]:
Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity which is essential for its chaperone activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function (PubMed:11274138, PubMed:15577939, PubMed:15937123, PubMed:27353360, PubMed:29127155). Engages with a range of client protein classes via its interaction with various co-chaperone proteins or complexes, that act as adapters, simultaneously able to interact with the specific client and the central chaperone itself (PubMed:29127155). Recruitment of ATP and co-chaperone followed by client protein forms a functional chaperone. After the completion of the chaperoning process, properly folded client protein and co-chaperone leave HSP90 in an ADP-bound partially open conformation and finally, ADP is released from HSP90 which acquires an open conformation for the next cycle (PubMed:27295069, PubMed:26991466). Apart from its chaperone activity, it also plays a role in the regulation of the transcription machinery. HSP90 and its co-chaperones modulate transcription at least at three different levels (PubMed:25973397). In the first place, they alter the steady-state levels of certain transcription factors in response to various physiological cues(PubMed:25973397). Second, they modulate the activity of certain epigenetic modifiers, such as histone deacetylases or DNA methyl transferases, and thereby respond to the change in the environment (PubMed:25973397). Third, they participate in the eviction of histones from the promoter region of certain genes and thereby turn on gene expression (PubMed:25973397). Binds bacterial lipopolysaccharide (LPS) and mediates LPS-induced inflammatory response, including TNF secretion by monocytes (PubMed:11276205). Antagonizes STUB1-mediated inhibition of TGF-beta signaling via inhibition of STUB1-mediated SMAD3 ubiquitination and degradation (PubMed:24613385). {ECO:0000269|PubMed:11274138, ECO:0000269|PubMed:11276205, ECO:0000269|PubMed:15577939, ECO:0000269|PubMed:15937123, ECO:0000269|PubMed:24613385, ECO:0000269|PubMed:27353360, ECO:0000269|PubMed:29127155, ECO:0000303|PubMed:25973397, ECO:0000303|PubMed:26991466, ECO:0000303|PubMed:27295069}.
Metal Binding:
N/A
Site:
N/A
Tissue Specificity:
N/A
Disease:
N/A
Mutagenesis:
MUTAGEN 47 47 E->A: Strong ATP-binding. Strong interaction with HSF1, HIF1A, ERBB2, MET, KEAP1 and RHOBTB2. No effect on interaction with TSC1. {ECO:0000269|PubMed:26517842, ECO:0000269|PubMed:29127155}.; MUTAGEN 93 93 D->A: Impaired ATP-binding. Strong interaction with HIF1A, MET, KEAP1 and RHOBTB2. Loss of interaction with HSF1 and ERBB2. Los of interaction with TSC1. {ECO:0000269|PubMed:26517842, ECO:0000269|PubMed:29127155}.; MUTAGEN 97 97 G->D: Abolishes ATPase activity. {ECO:0000269|PubMed:18256191}.; MUTAGEN 313 313 Y->E: Loss of interaction with TSC1 and increases interacrion with AHSA1. {ECO:0000269|PubMed:29127155}.; MUTAGEN 313 313 Y->F: No deffect on the interaction with TSC1. {ECO:0000269|PubMed:29127155}.; MUTAGEN 598 598 C->A,N,D: Reduces ATPase activity and client protein activation. {ECO:0000269|PubMed:15937123, ECO:0000269|PubMed:19696785}.; MUTAGEN 598 598 C->S: Loss of S-nitrosylation. {ECO:0000269|PubMed:15937123, ECO:0000269|PubMed:19696785}.; MUTAGEN 728 732 Missing: Loss of interaction with TSC1. {ECO:0000269|PubMed:29127155}.
Reagent Data
Name:
Heat shock protein HSP 90-alpha (Heat shock 86 kDa) (HSP 86) (HSP86) (Lipopolysaccharide-associated protein 2) (LAP-2) (LPS-associated protein 2) (Renal carcinoma antigen NY-REN-38)
Class:
Subcategory:
Recombinant
Molecular Weight:
Source:
Species:
Human
Amino Acid Sequence:
Tag:
Format:
Solution
Formulation:
Sterile-filtered colorless solution
Formulation Concentration:
1mg/ml
Buffer Volume:
20mM
Buffer Solution:
Tris-HCl
pH:
7.4
Stabilizers
NaCl:
100mM
Metal Chelating Agents
EDTA:
Null
Purity:
> 90%
Determined:
SDS-PAGE
Stained:
Assay (Variable)
Validated:
Inquire
Sample Handling
Storage:
4°C
Stability:
This bioreagent is stable at 4°C (short-term) and -70°C(long-term). After reconstitution, sample may be stored at 4°C for 2-7 days and below -18°C for future use.
Preparation:
Reconstitute in sterile distilled H2O to no less than 100ug/ml; dilute reconstituted stock further in other aqueous solutions if needed. Please review COA for lot-specific instructions. Final measurements should be determined by the end-user for optimal performance.